-
Tcl, Standard File Open/app Launch Commands For Mac카테고리 없음 2020. 1. 31. 05:28

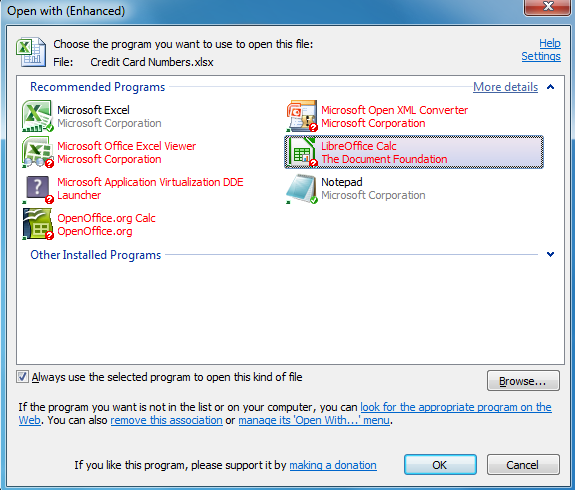
Tcl Standard File Open/app Launch Commands For Mac Free
When you open VMD, by default a vmd console window appears. The vmd console window tells you what's going on within the VMD session that you are working on. 2 Take a look at the vmd console window (Fig. ). It should tell you a molecule has been loaded, as well as some of its basic properties like number of atoms, bonds, residues and etc. The Tcl commands that you enter in the VMD TkConsole window can also be entered in the vmd console window.
Support files, caches, and app-generated data files should be placed in one of the standard directories dedicated to app-specific files. For information on how to present and customize the Open and Save panels, see Using the Open and Save Panels. Once the download is completed, you can navigate to your Downloads and double-click on the disk image file. This will mount the image and when it opens, you can control-click on the dot mpkg file. This will give you a menu where you can choose to open the file. Selecting Open will bring up the Python Installer. Click Continue to progress through the menus. Project Participants. GPG/PGP keys of package maintainers can be downloaded from here. If you would like to see a map of the world showing the location of many maintainers, take a look at the World Map of Debian Developers.
If you are using a Mac, your vmd console window is the terminal window that shows up when you open VMD. Atom selections are just references to the atoms in the original molecule. When you change a property (e.g. beta value) of some atoms through a selection, that change is reflected in all the other selections that contain those atoms. 8 In the Tk Console window, type set sel atomselect top 'hydrophobic'. This creates a selection, sel, that contains all the atoms in the hydrophobic residues. 9 Let's label all hydrophobic atoms by setting their beta values to 1.
You probably know how to do this by now: type $sel set beta 1 in the Tk Console window. If the colors in the OpenGL Display do not get updated, go to the Graphical Representations window and click on the Apply button at the bottom. 10 You will now change a physical property of the atoms to further illustrate the distribution of hydrophobic residues.
Launch Commands For Pubg
In the Tk Console window type $crystal set radius 1.0 to make all the atoms smaller and easier to see through, and then $sel set radius 1.5 to make atoms in the hydrophobic residues larger. The radius field affects the way that some representations (e.g., VDW, CPK) are drawn. You have now created a visual state that clearly distinguishes which parts of the protein are hydrophobic and which are hydrophilic. If you have followed the instructions correctly, your protein should resemble Fig.
